Input for the CysDuF Database :
The input for the CysDuF database can be a DUF ID, PDB ID or an PFAM ID. The input, DUF_ID or PDB_ID or the PFAM_ID should be relevant to these pathways namely ElectronTransportChain, Glutathione Metabolism, Fe-S-Cluster Biogenesis, Fatty Acid Synthesis, Photosynthesis, Krebs Cycle and Pentose Phosphate Pathway.The output can be printed in a separate webpage or also can be downloaded as a CSV file, JSON file or an TXT file.
Output for the CysDuF Database :
- PFAM ID : The PFAM ID is from the Pfam database where it is an secondary database and is an widely used database. The database is a widely used web resource for the classification of protein sequences into families and domains.
- DUF ID : DUF ID is the Domains of Unknown Function ID. Domains of Unknown Function is the protein domains where the protein function is unknown.E.g., DUF4563
- DUF_Name : The name of the DUF protein is mentioned where it can be for E.g. Domain of the Unknown Function (DUF4563)
- Name of the DUF : The name of the DUF is the name that belongs to the SCOPe database family and using biological and the strucutral information of the respective DUF protein has been curated.
- Species: The species information is retrieved from the SCOPe database and the organism can belong to any of the four classification kingdoms namely Bacteria, Virus , Archeabacteria and Fungi . The classification is based on the NCBI taxanomy classification , and these organisms can cause different kinds of diseases. Majority of the diseases are caused by the Bacteria based on the data statistics. Coronavirus has been firstly reported in the DUFs proteins and few plant diseases has also been reported
- SCOPe Superfamily and Family: SCOPe database is an extended database for the strucutral classification of the proteins which explains the proteins strucutre and evolutionary releationships. The SCOPe database is classified into Class, Fold, Superfamily, Family, and the Superfamily defines the proteins that originate from a ancestor and has common strucutral and functional features. Family information of the SCOPe database is also given in this database where it defines the proteins with similar sequences but the functions are distinct.

SCOPe hierarchy
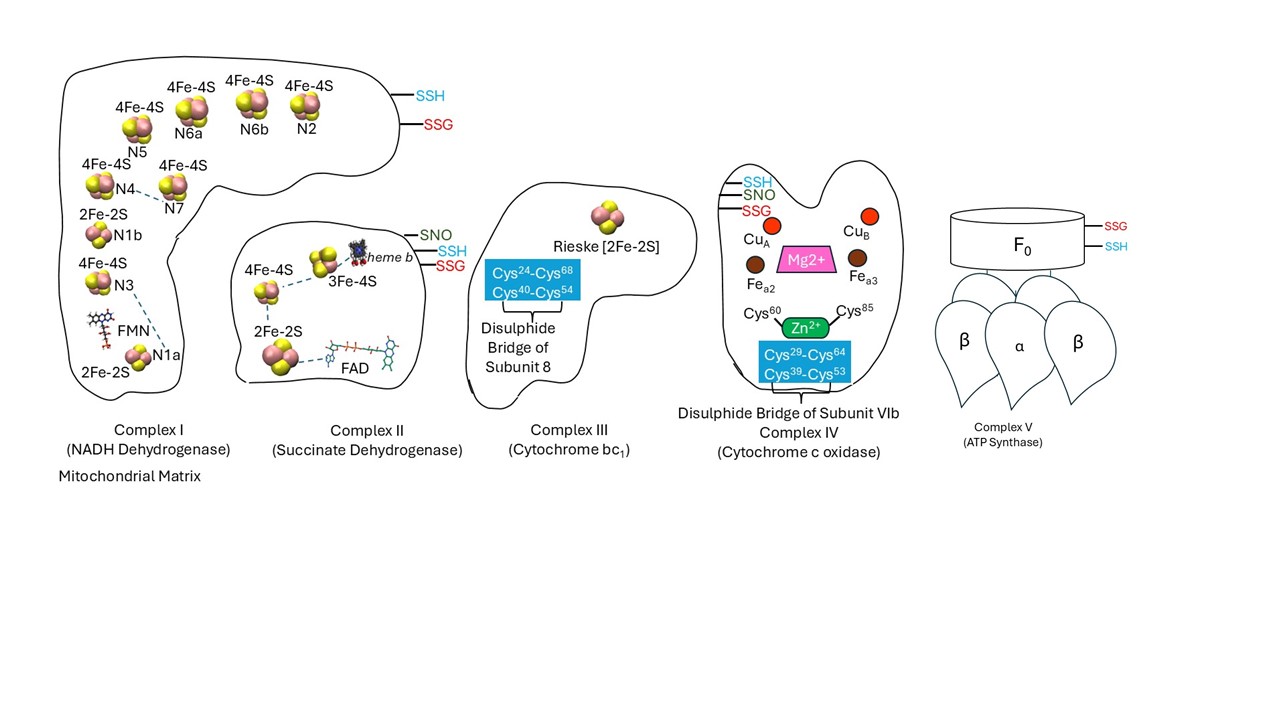
- Pathway: In this it gives information to which pathway that the DUFs proteins belongs to . In this database it is only focused on seven pathways namely ElectronTransportChain, Glutathione Metabolism, Fe-S-Cluster Biogenesis, Fatty Acid Synthesis, Photosynthesis, Krebs Cycle and Pentose Phosphate Pathway.The DUFs proteins belonging to these pathways are only present in this CysDuF database.
- PDB_ID and Chain_ID: The PDB and Chain_ID information is curated from PDB Database of the respective PDB file.
- DeepCys Predictions: Cysteine residues are annotated for the four Cysteine post-translational modifications namely Disulphide, S-Sulfenylation, Metal-binding and Thioether based on the DeepCys . DeepCys predictions is an multiple Cysteine function prediction algorithm based on the deep neural network and was trained and tested on two independent datasets curated from protein crystal strucutres.
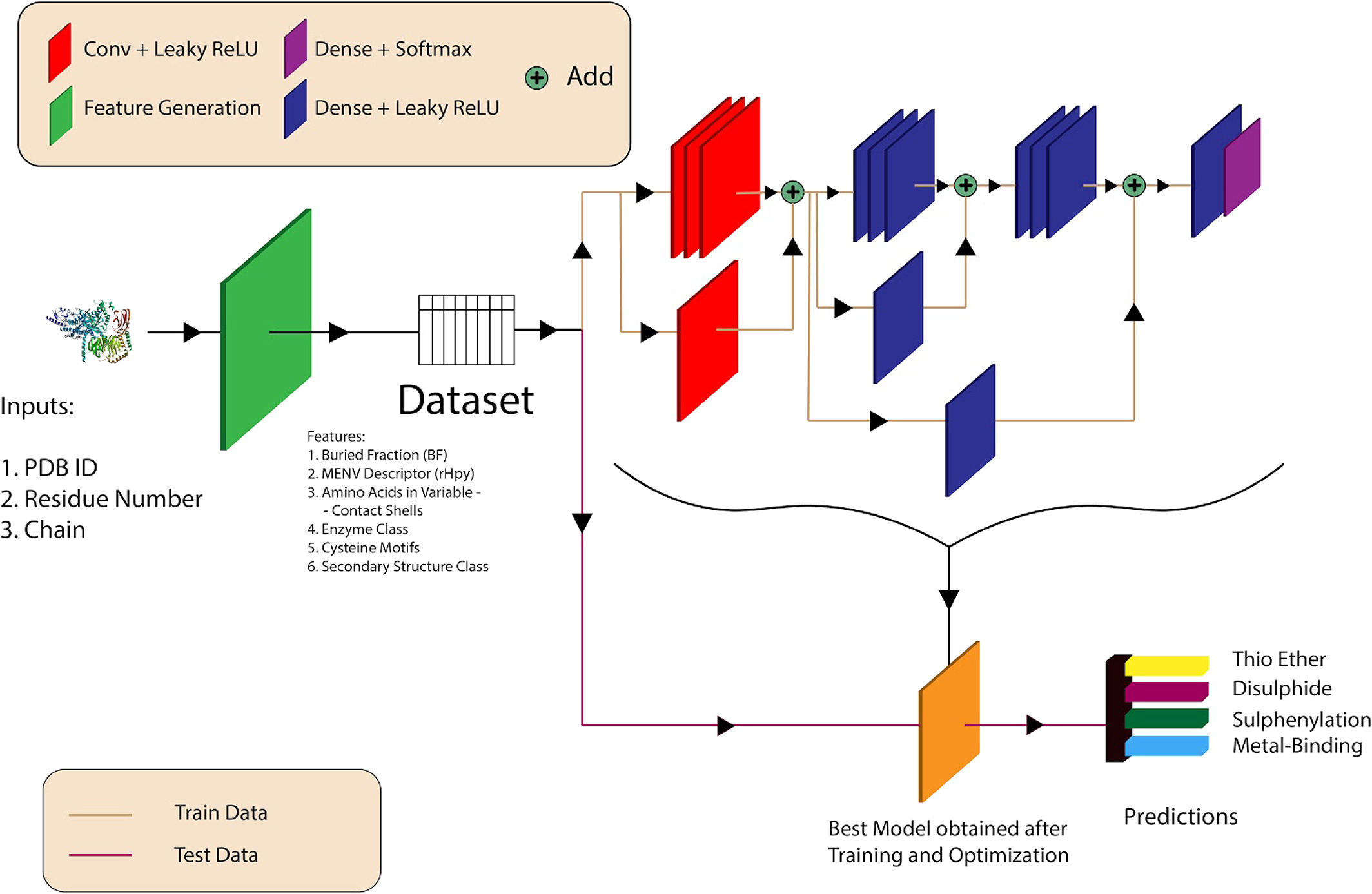
Workflow and architecture of the deep learning model for structure-based prediction
- Buried Fraction: It is an component of the MENV protein microenvironment computed for the given PDB strucutre and is described as the normalized surface area of cysteine thiol group buried inside the protein. Buried Fraction parameter of the protein ranges from 0.0 to 1.0 and zero buried fraction indicates that the thiol group is completely exposed to the solvent.
- Relative Hydrophobicity (rHpy): It is another MENV protein microenvironment property descriptor describes the relative hydrophobic contribution of the protein and solvent towards the Cysteine thiol group within its first contact shell.