1.) The input for the CysDuF database webserver can be an DUF ID, Pfam ID, Species or an PDB ID. The Pfam ID is from the Pfam database1 which is a secondary database and consists of the protein families and domains. PDB ID is retrieved from the PDB database2 which is a primary database consists of the protein strucutres.
 2.) The results can be downloaded in form of a CSV file, JSON file or an TXT file.
2.) The results can be downloaded in form of a CSV file, JSON file or an TXT file.
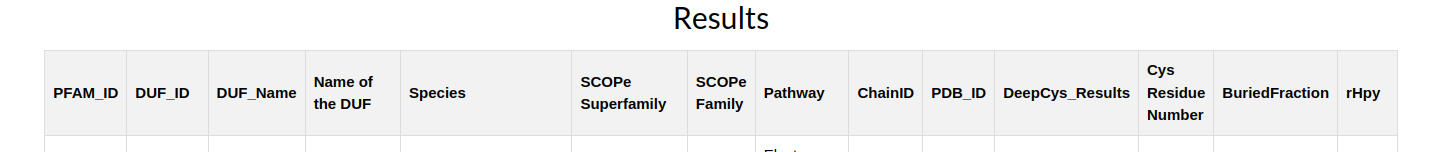
 3.) Results are also printed in a separate web page in form of an tabular column where it consists of a Pfam ID , retrieved from the Pfam database1, DUF information with the DUF ID and DUF Name, Species information of the PDB ID retrieved from the SCOPe database3, and the SCOPe database information namely the SCOPe Superfamily , SCOPe family information, Pathway associated with the DUFs protein, PDB_ID, Chain_ID retrieved from the SCOPe database where as the Cysteine Residue Number is retrieved from the PDB database2, DeepCys Predictions4 are prediction results from the DeepCys Cysteine multiple Cysteine post-translational modifications strucutre based prediction and the Buried fraction and Relative hydrophobicity (rHpy) is retrieved from the MENV5.
3.) Results are also printed in a separate web page in form of an tabular column where it consists of a Pfam ID , retrieved from the Pfam database1, DUF information with the DUF ID and DUF Name, Species information of the PDB ID retrieved from the SCOPe database3, and the SCOPe database information namely the SCOPe Superfamily , SCOPe family information, Pathway associated with the DUFs protein, PDB_ID, Chain_ID retrieved from the SCOPe database where as the Cysteine Residue Number is retrieved from the PDB database2, DeepCys Predictions4 are prediction results from the DeepCys Cysteine multiple Cysteine post-translational modifications strucutre based prediction and the Buried fraction and Relative hydrophobicity (rHpy) is retrieved from the MENV5.